(Clinical) Therapeutic
Antibody Database
Therapeutic antibody database extended with clinical information, with special focus on immunogenicity annotations.
Learn moreIntroduction
Therapeutic antibody data is crucial for characterization of novel biologics therapeutics. Current therapeutic antibody data such as format, sequence and clinical data are spread across different repositories. Cleaning this data allows us to forecast developability and clinical trial outcomes based on the antibody sequence and the said metadata.
In order to introduce organization to this data, we created our Therapeutic Antibodies Database, that introduces a format which seamlessly associates the different metadata with any target therapeutic and is available for download and computational/machine learning applications. The biggest advantage of the format of our dataset is that it is easily extensible as novel information on therapeutics is collected. We demonstrate the flexibility of the novel format by associating the Anti-Drug Antibody data via a comprehensive review of clinical data points on immunogenicity.
The therapeutic database contains full sequence information, post translational modifications and target annotations for 1 109 therapeutics. The therapeutics were assigned to 117 distinct formats, categorized into 5 groups(Figure 1). We have identified 438 unique targets, all normalized and extended with UniProt data.
1 109
Therapeutics
438
Targets
117
Formats categorized into 5 groups
1 138
Clinical trials measuring
immunogenicity
~12 000
Anti-Drug Antibody
points
Data availability & access
We make the dataset available to non-profit organizations for non-commercial purposes. Other organizations & usage types, are kindly asked to contact us via contact us.
Category Formats present in the database:
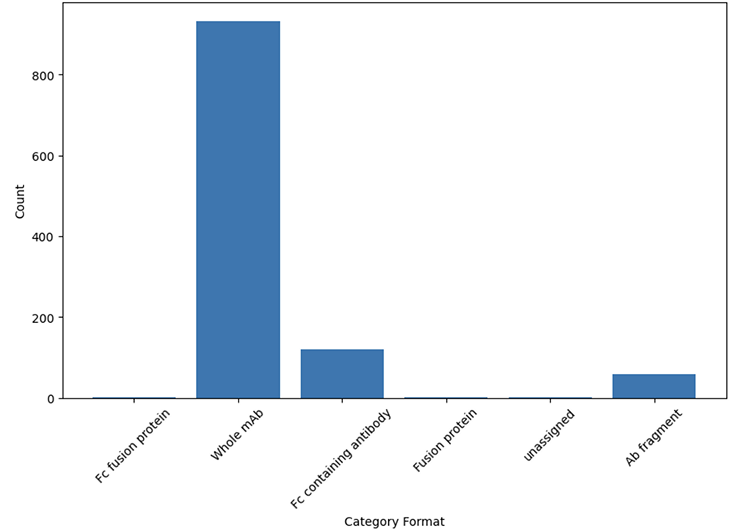
Figure 1. Therapeutic format categories present in the database
For all of the therapeutics from the database, we have searched clinicaltrials.gov using the names and synonyms. This yielded 26 996 therapeutics-related clinical trials, measuring distinct outcomes. We have identified clinical trials in which immunogenicity was measured using our in-house semantic search engine. This yielded 1 138 clinical trials measuring immunogenicity data in 4 398 cohorts with the overall number of identified measurements equal to ca 12 000, which are all present in the database. The measurements cover ~300 distinct therapeutics.
There are a total 277 unique diseases assigned to all cohorts, which we grouped into 10 categories (Figure 2).
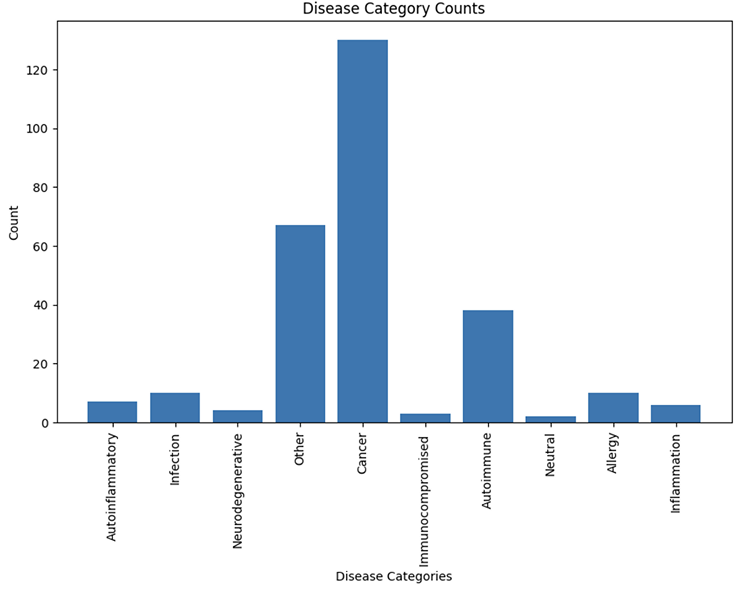
Figure 2. Distribution of disease categories assigned to cohorts.
We have identified a total of 7146 cohort interventions and assigned the dosages and routes of administration to them (Figure 3).
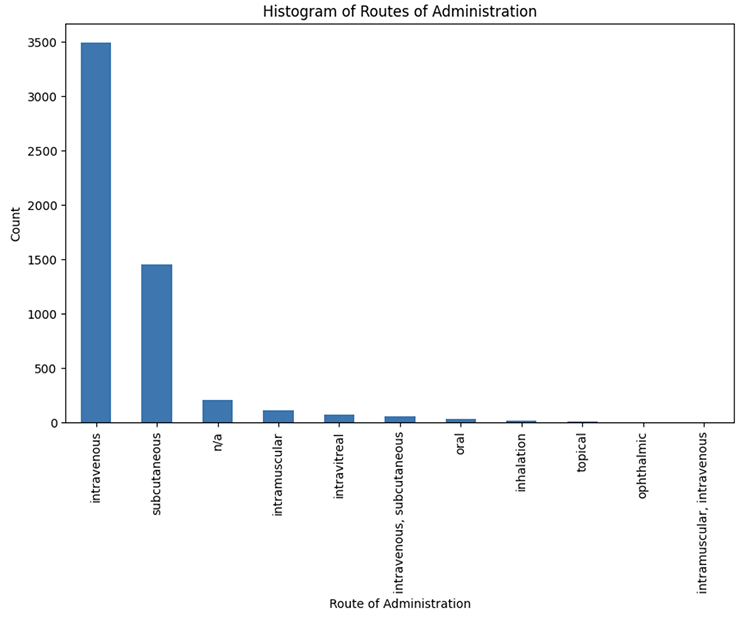
Figure 3. Distribution of routes of administration for all interventions
We have harmonized the measurements, as described in the annotation pipeline to produce the subset datasets, representing incidence, prevalence, and pre-existing immunogenicity, consisting of
801, 2274, 576 records respectively.
Data format
The data is formatted as .csv into several sheets, akin to excel. This was dictated by the fact that a single therapeutic needs to be associated with multiple values in a relational database fashion, which is not suitable for a single .csv file. Sharing dumps of relational databases though is not user friendly. The entity relationships diagram of the database tables is presented on Figure 4.
Figure 4. Entity relationship diagram of the database tables.
At the core of the database is a therapeutic table, which contains drug name, its synonyms, information about its format, and postranslational modifications. Each therapeutic is sourced from inn lists, so references to those are stored in the inn_lists.
Each therapeutic consists of one or more amino acid chains, which are stored in the molecule table, together with their full sequences, and target antigens. Each molecule may contain variable domains, which are stored in the variable_domain table, together with the position of the domain in the sequence.
So for example a typical monoclonal antibody which is built from heavy and light chains will have 1 record in the therapeutic table, 2 records in the molecules table and 2 records in the variable_domain table (one assigned to each molecule). On the other hand, a scFv therapeutic will have only one molecule with multiple variable regions assigned to it.
Each therapeutic may have some clinical trials assigned to it in which it was tested. The table clinical_trial contains basic trials information, such as title and phase. As the relationship between therapeutic table and clinical_trial table is many to many, it is realized with a third table holding references from both tables (therapeutic name and clinical trial identifier).
Each clinical trial measures various outcomes. We focus on the outcomes investigating immunogenicity. The immunogenicity may be measured at multiple points in time, so each distinct point in time should have a separate record in the outcomes table. The immunogenicity in clinical trials is measured for each cohort separately. Each cohort will have associated diseases and interventions in the corresponding tables. Interventions table contains information about the treatment regimens if they are present, including the dosage and the route of administration.
Finally the measurement table contains immunogenicity measurements associated with corresponding clinical trials outcomes, and cohorts. The values were properly annotated to facilitate inter- study comparisons. For a more detailed description, see the following chapter.
From all of the above tables we derive 3 datasets presenting incidence, prevalence and pre-existing immunogenicity.
Curation of the database
We manually review INN lists, published by WHO to identify antibody therapeutics and extract full sequences (including constant regions), target information, and post-translational modifications. Molecules are numbered via the IMGT scheme using RIOT, with variable domains described in the AIRR Format. Trade names are imported from OpenFDA, synonyms from the NCI Thesaurus.
Each therapeutic is defined as a “Therapeutic” entity comprising one or more amino acid chains (“Molecule” records). Molecules with defined targets receive curated Uniprot IDs and sequences; post-translational modifications are captured directly from INN lists.
A database of antibody immunogenicity measurements was constructed by compiling monoclonal antibody names and synonyms, then importing relevant clinical trials from ClinicalTrials.gov into tables such as clinical_trial, outcome, result_group, measurement, disease, and interventions. Clinical trials report multiple outcomes (toxicity, pharmacodynamics, immunogenicity) at specific time points per cohort (result_group). We focus only on immunogenicity related outcomes.
Cohorts were annotated with diseases and interventions, so that each cohort has diseases assigned, and treatment interventions, together with dosages and routes of administration (if present).
Given the heterogeneity in how studies reported their results, several strategies were implemented to standardize the data. When multiple measurements for a single outcome were reported at different time points, artificial outcomes were created so that each outcome corresponded to a single time point. Neutralizing antibody responses were specifically flagged, allowing for their separation from other immunogenicity measurements. In instances where partial results were reported, a grouping key was assigned to facilitate proper aggregation. An example of partial measurements are measurements of immunogenicity at different titer levels or performing more accurate measurements after the screening.
Each measurement was annotated with a type indicator to distinguish between positive, negative, missing or inconclusive outcomes. Additionally, studies that reported overall immunogenicity alongside relative measures - such as treatment-emergent or treatment-boosted responses - were annotated accordingly. This approach enabled direct comparisons, with overall immunogenicity defined as the sum of pre-existing, treatment-boosted and treatment-emergent responses.
Further data harmonization included adjusting measurement group sizes where the number of eligible patients differed from the initially defined cohort size. In some cases eligible patients were extracted from metadata; in other cases they were extracted from the measurement description; in the remaining cases eligible patients were derived by subtracting missing and inconclusive participants from the original cohort size. This unification allowed for another unit conversion - measurement units were normalized to represent the percentage of participants, ensuring consistency across studies. In cases where studies differentiated between transient and persistent measurements, transient responses (defined as positive at one time point and negative at another) were flagged and annotated with a grouping key to allow for correct summation with overall ADA scores.
With the above annotations we constructed 3 additional derived datasets: presenting immunogenicity incidence, prevalence and baseline for cohorts in which only one antibody therapeutic was administered.
We define immunogenicity incidence as the presence of treatment emergent or boosted ADA so that only relative measurements are considered, performed after the drug administration. Treatment emergent measurement is defined as the number of ADA positive participants (or percentage of participants) who were ADA negative at baseline (before the drug administration). Similarly, treatment boosted measurements are defined as positive post baseline participants, who were also positive before the drug administration, and there was a significant change in titer levels (usually 4-fold).
Immunogenicity prevalence is defined as the measurements of all patients without the stratification on the baseline results. In other words the prevalence represents incidence, but together with baseline positive measurements.
Finally the baseline dataset was constructed to investigate the pre existing immunogenicity of the therapeutics. To build this subset, we gathered all of the measurements from placebo groups and all groups in which the ADA was measured before the drug administration.
For all three datasets, after we grouped partial measurements and unified the units (participants were converted to percentage values based on identified groups sizes), we grouped the measurements from multiple points in time, so that each cohort had.
Data availability & access
We make the dataset available to non-profit organizations for non-commercial purposes. Other organizations & usage types, are kindly asked to contact us via contact us.
Citing this work
We make the Therapeutic Antibody Database available as a companion to our paper, that will be posted here once it clears peer review.
Therapeutic Antibody Database - repository of clinical antibody information with associated clinical data, with special focus on immunogenicity.