INDI2 Database
Integrated Nanobody® Database for Immunoinformatics, Version 2.0. The curated repository of single-domain antibody data, powering the next generation of immunoinformatics and generative protein design.
Introduction
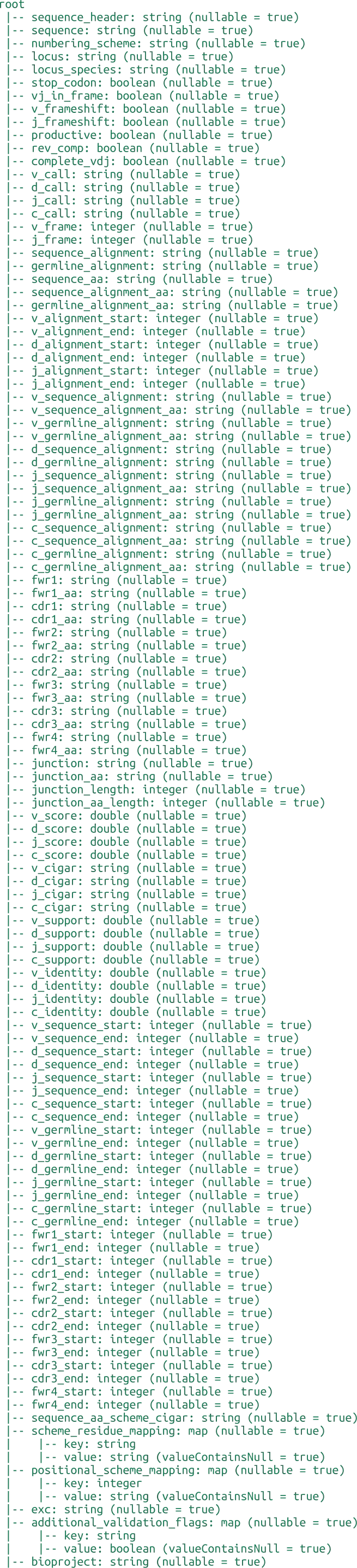
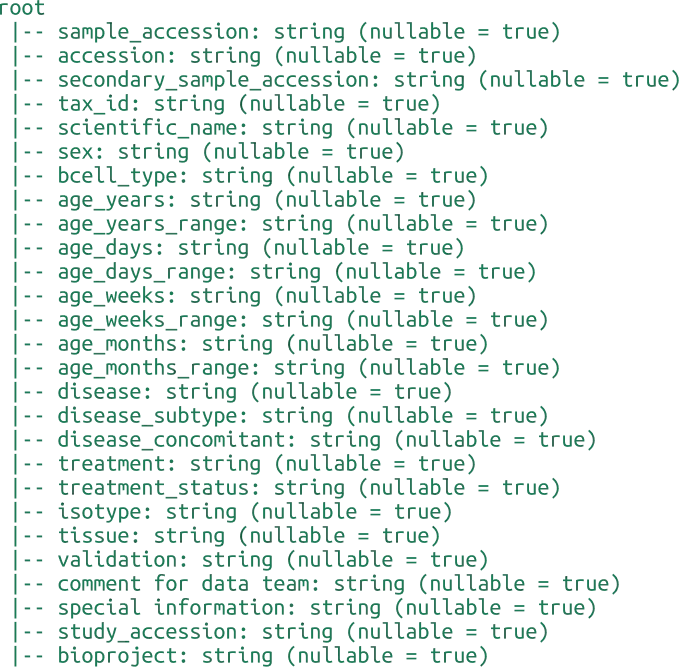
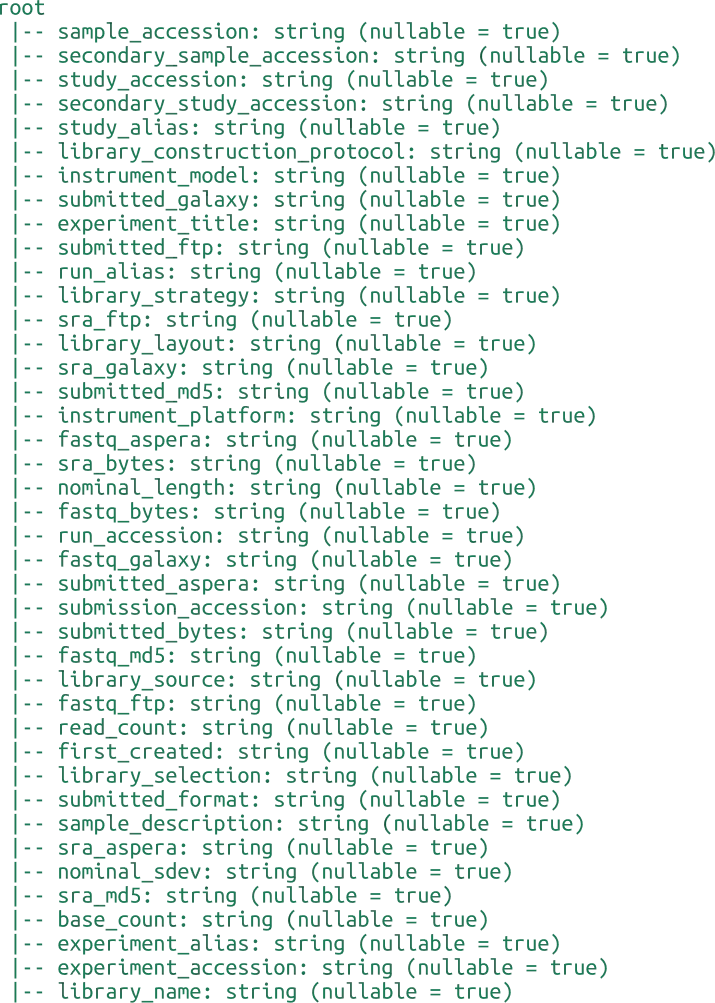
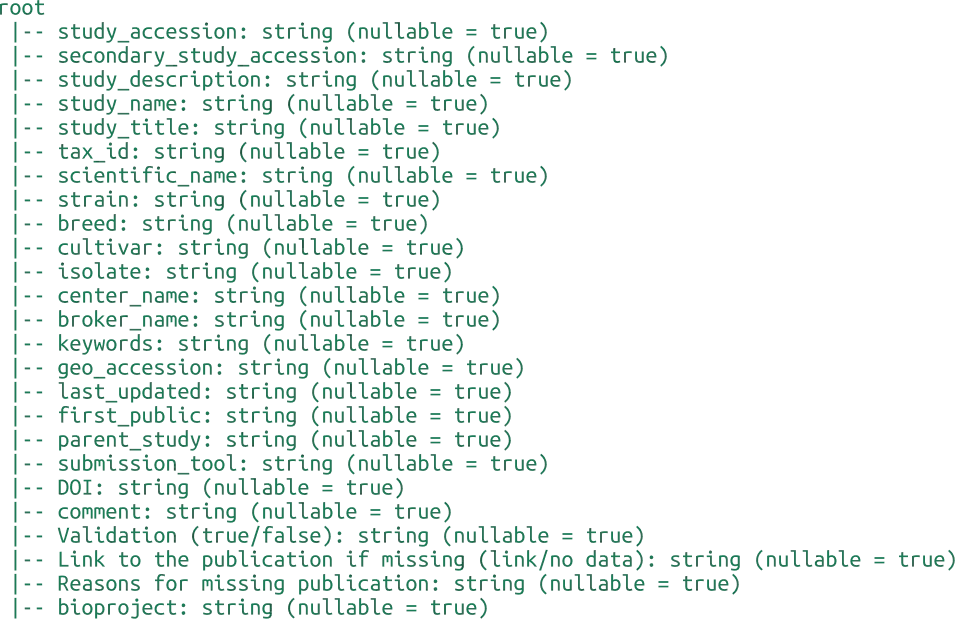
INDI2 DB is a comprehensive repository of Nanobodies® (VHH) sequences, structures, and metadata designed to support computational research in molecular modeling, protein engineering, and machine learning. Unlike general-purpose repositories, INDI2 is a curated ecosystem that aggregates and harmonizes data from six distinct pillars of the NaturalAntibody infrastructure.
The current version of the database is a strict subset of sequences and structures derived from NaturalAntibody’s specialized databases: the Patents Database, NGS Database, Structural Database, Therapeutic Antibody Database, Antigen-Specific Antibody Database (AgAb), and GenBank® Database.
Nanobodies® (VHH) selection engine
We have applied three distinct heuristics to detect Nanobody® sequences:
Assigned alpaca germline - whether or not RIOT assigned alpaca germline
Camelidae family and Sharks - whether the sequence originated from Camelidae family in taxonomy tree or were derived from Sharks
GPT like (set of various models like Gemini 2.5 Flash, GPT o3-mini etc. is being validated and applied during the pipeline execution so it`s changing over time) annotation indicating the publication corresponding to GenBank record or Patent description relates to Nanobodies®
Flags should be considered in combination, as relying solely on germline assignment might lead to inaccuracies as detection by germline might falsely positively flag sequences not originating from camelids. On the other hand, synthetic sequences are not detected with the by_family flag. GPT annotation constitutes the smallest contribution to the dataset. Each record contains the Nanobody® markup source giving also the possibility to make more restricted heuristic if required.
Nanobodies® from the NA Structural DB were marked based on the lack of correct pairing of the Heavy Chain to the corresponding Light Chain with chains detection using RIOT in the first place.
6
Number of data sources
~30M
Nanobody® sequences
Specific database amount of records
Database
Records
Definition
NAStructural DB
- 2 768 Nanobody® -antigen complexes, including a curated subset of 799 representative structures derived from nanobody and antigen clustering
- 4 160 unbound (single-chain) Nanobody® structures, with a refined subset of 884 representatives identified through Nanobody® clustering
Patents DB
67 479 Nanobody® sequences extracted from 1 919 patent families. With 18 911 sequences identified and mapped to their specifically assigned targets.
Therapeutic Antibodies DB
15 therapeutics constructed with Nanobody® molecules cross-referenced with 175 available clinical trial records.
GenBank®
4 169 unique (non-patent) Nanobody® GenBank® sequences from 4 145 accessions.
ASD
110 935 Nanobody®-antigen records enriched with binding data. These measurements were compiled from the Antigen-Specific Antibody Database (ASD)
Table 1. Statistics of INDI2 database. We sourced Nanobody® sequences from internal NaturalAntibody datasets. We present the total number of sequences per dataset - these might be redundant within and across the sources. Since metadata such as target annotations are not universally present across all sources, we left those out from these statistics.
Accessing the Database
INDI2 DB is freely available for non-commercial organisations for non-commercial research. Commercial inquiries are welcome via contact us.
Directory structure

INDI2 is accessible using google drive with the following directory structure. It allows you to download a single dataset in specific format or all datasets at once.
Data format
The INDI2 dataset is available in parquet/delta (delta.io) format streamlining the integration into data processing pipelines (using Apache Spark, Pandas, DuckDB).

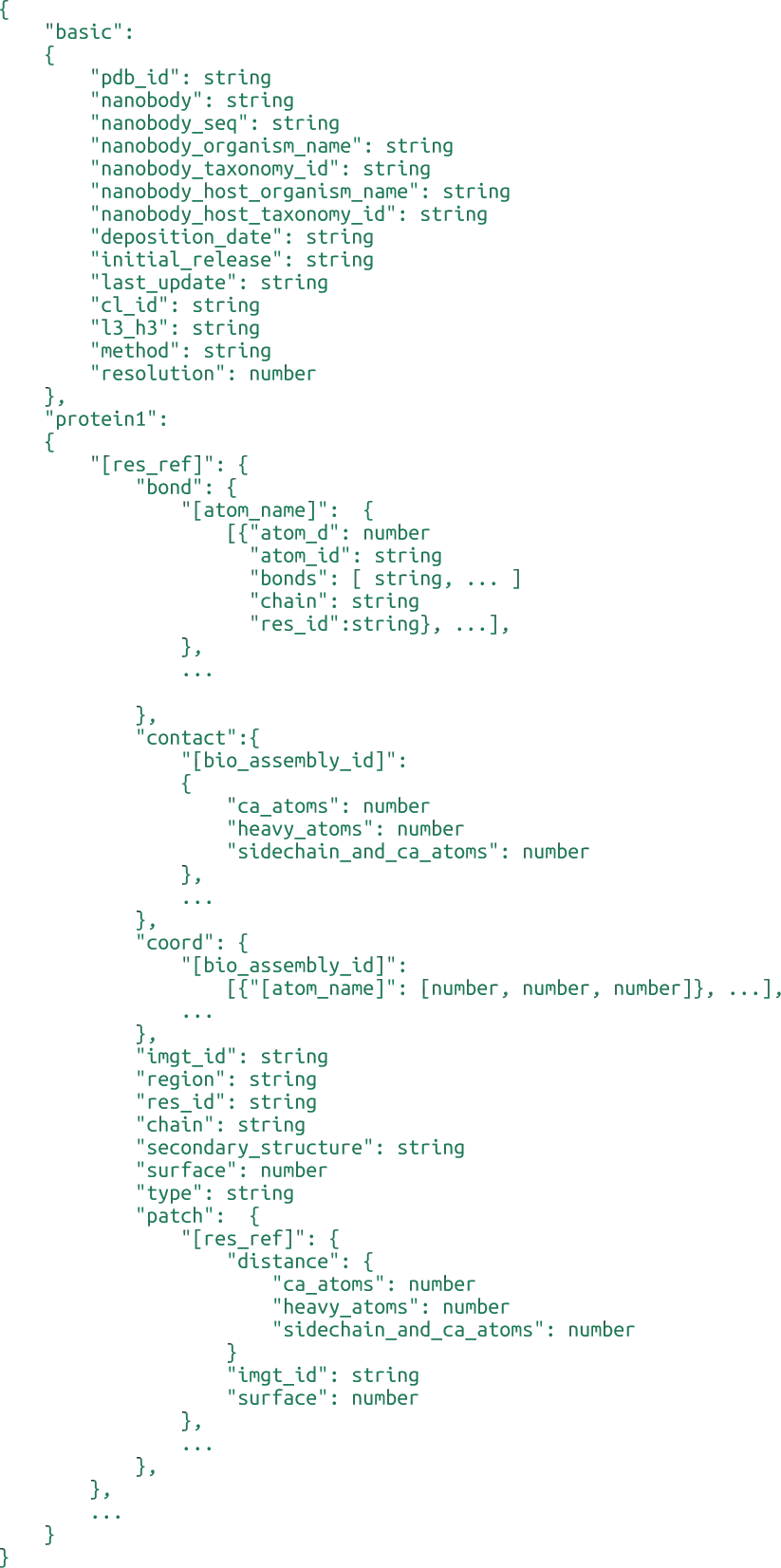
Patents Database
Source Database Details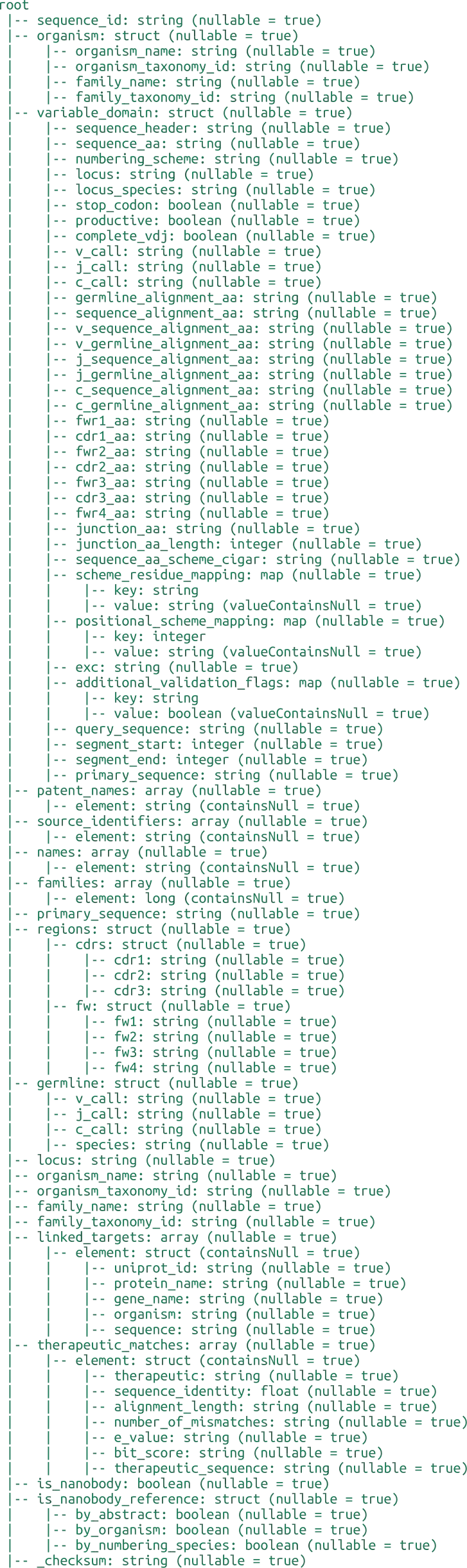
Antigen Specific Antibody Database
Source Database Details
INDI - NANOBODY® DB - 2021
INDI - Integrated NANOBODY® Database for Immunoinformatics. Automatically collected NANOBODIES® (or single domain antibodies, VHH) from heterogenous public sources. It covers Structures, Patents, GenBank®, Next Generation Sequencing Repositories.
17129 unique NANOBODY® patent-sequences from 843 patents.
946 unique NANOBODY® structure-sequences from 1587 PDBs.
1950 unique (non-patent) NANOBODY® GenBank®-sequences from 2179 accessions.
11228600 NGS unique NANOBODY® sequences from 7 bioprojects.
1268 sequences manually curated from 109 publications.
Table 2. Statistics of INDI Database.
Accessing the Database
We make the INDI2 DB free for non-commercial use by non-commercial entities. If you are a commercial entity and would like to employ the data in your activities, please get in touch with us.
For additional support or to report any issues, please reach out via our website
Citing this work
If you use either INDI1 or INDI2, please cite this paper:
INDI—Integrated Nanobody® Database for Immunoinformatics
Authors: Piotr Deszyński, Jakub Młokosiewicz, Adam Volanakis, Igor Jaszczyszyn, Natalie Castellana, Stefano Bonissone, Rajkumar Ganesan, Konrad Krawczyk
Journal: Nucleic Acids Research, Volume 50, Issue D1, 7 January 2022, Pages D1273-D1281, https://doi.org/10.1093/nar/gkab1021