Antigen Specific
Antibody Database
Comprehensive repository supporting computational research in antibody engineering and therapeutic design, with a focus on antibody-antigen interactions
Introduction
AgAb DB (Antigen Specific Antibody Database) is a comprehensive repository supporting computational research in antibody engineering and therapeutic design, with a focus on antibody-antigen interactions. This database addresses the challenge of dispersed binding data by aggregating and standardizing antibody-antigen interaction information from multiple sources.
AgAb DB is freely available for non-commercial research use. The database can be accessed through the Download section, where its files can be downloaded from Google Drive, or via the Google Colab, which provides a ready-to-use notebook with step-by-step guidance for loading and working with the database directly in Colab. Colab notebook showcases the basics of downloading, loading, and filtering of the database, along with the necessary dependencies to make it happen. The overview of the dataset statistics, schema, and dataset composition is presented in the section below.
Data sources
The primary dataset sources creating the database:
1. Genbank Database
2. SKEMPI 2.0
3. Peer-reviewed publications
4. Patent datasets
The statistics describing the dataset do not include antibodies targeting HER2 antigen, due to its large share in the dataset (524 346 samples) and patents (113 117 samples) due to its moderate confidence.
457 889
Total Antibody-Antigen
Records
309 884
Unique Antibodies
170 660
Complete Heavy/Light
Chain Pairs
4 334
Unique Antigens
Antigen Distribution: Primary focus on infectious diseases and cancer targets
Affinity Measurement Types: Includes quantitative metrics (Gibbs free energy changes, kinetic constants, IC₅₀) and qualitative binding assessments
Antibody Structure: The majority of entries include both heavy and light chains
Sequence Length Distribution:Predominantly in the "short" category (<150 amino acids)
The dataset contains 70 388 nanobodies among non-patent databases (113 380 with patents) and 132 157 scFv antibodies.
Nanobodies:
Alphaseq: 67 058 entries
Patents: 40 517 entries
Literature: 1 936 entries
Structures: 1 258 entries
AATP: 93 entries
OSH: 30 entries
RMNA: 10 entries
scFv:
Alphaseq: 131 645
Literature: 512
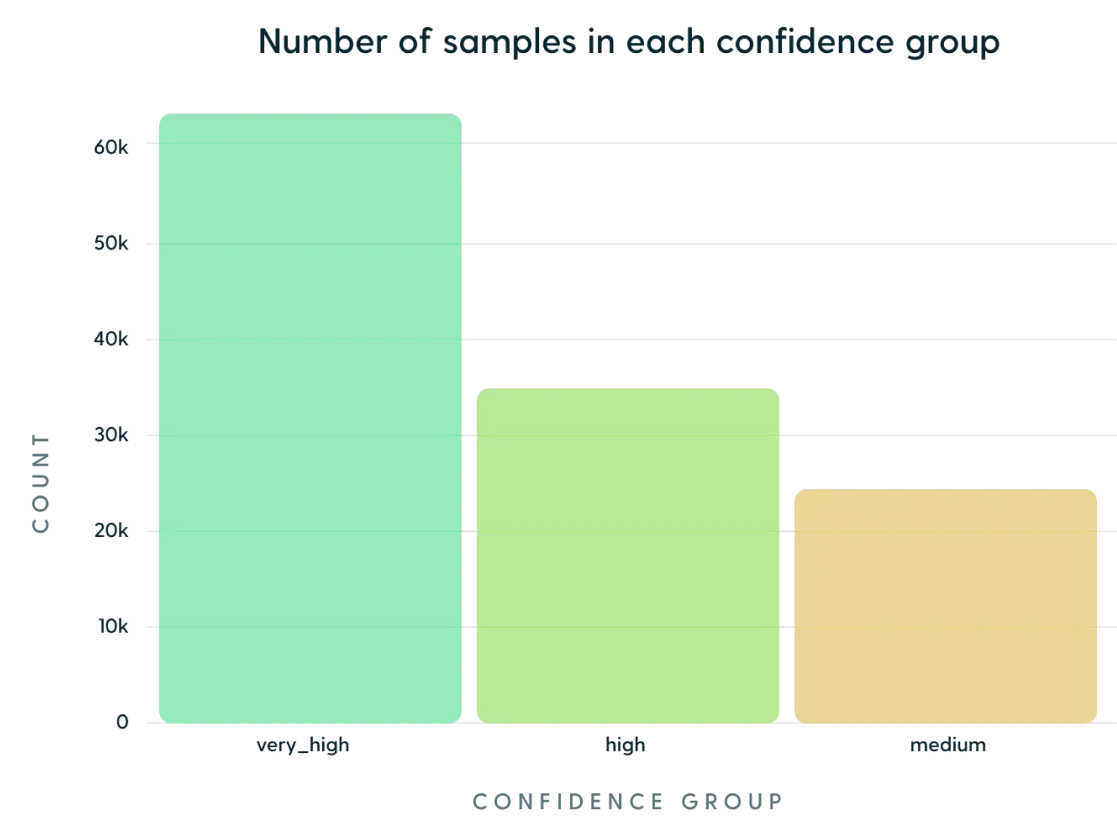
The dataset was split into 3 confidence categories: medium, high and very_high, with very_high being the most dominant (51%) group:
The “very high” category means that both the sequences and methodology used for calculating the affinity were robust, an example of such a dataset is AbDesign.
The “high” confidence represents datasets which have to be manually curated or contain only the name and/or mutations of the target antigens/antibodies. The flab dataset is a member of this group, as it includes antigen names, not their respective sequences.
“Medium” category is a product of automated data discovery, which, although validated, can contain a degree of uncertainty, like patents database.
Accessing the Database
AgAb DB is freely available for non-commercial organizations for non-commercial research. Commercial inquiries are welcome via contact us.
It can be accessed in a manner described in the colab notebook.
Statistics of each dataset can be seen below
Dataset
Is naturalantibody dataset
Record count
unique antigens
unique antibodies
affinity_type
confidence
literature
True
5580
874
4841
bool
high
Category Formats present in the database:
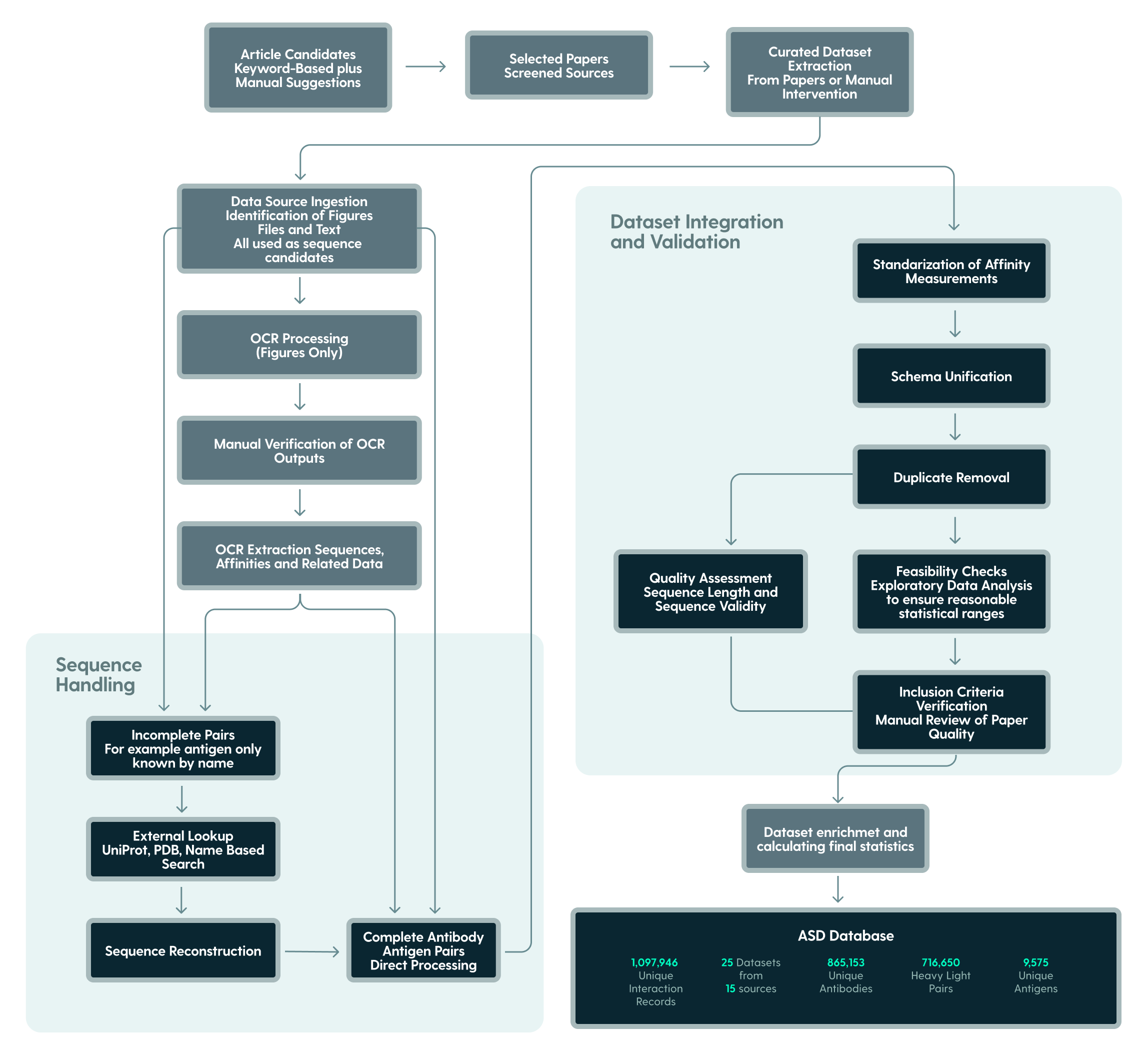
Data processing pipeline
1. Aggregation: Collection from 14 (and baselining the buzz, which was excluded in this description as it dominates other datasets in size) distinct sources resulting in 25 integrated datasets
2. Curation: Multi-stage pipeline combining automated extraction, normalization, and manual verification
3. Standardization: Common structure implemented across all studies
4. Validation: Automated feasibility checks and manual verification of critical datasets
Data structure
The dataset follows a standardized schema for all studies, containing Antibody-Antigen Pairs along with affinity measures:
Name
Description
Example
dataset
Dataset abbreviation that allows for reading about its characteristics in this documentation
DLGO
heavy_sequence
Heavy chain sequence of the antibody written using amino acids
QVQLVQSGAEVKKPGAS…
light_sequence
Light chain sequence of the antibody written using amino acids
DIQMTQSPSTLSASVGD…
affinity_type
Type of measurement perform to obtain binding affinity
ic_50
affinity
Numerical value obtained using method described in affinity_type
0.002
antigen_sequence
Antigen sequence expressed in amino acid format
MFVFLVLLPLVSSQCVN…
metadata.target_uniprot
Id of an antigen connecting it to a specific Uniprot database entry
P0DTC2
metadata.target_pdb
Id of an antigen connecting it to a specific PDB database entry
7EB0
metadata.target_name
Name or label for the antigen target (e.g., "covid_gamma").
covid_gamma
confidence
Level of confidence in binding of exact antibody sequence to an exact antigen sequence
high
scfv
Boolean value specifying whether records containing only heavy sequence contain a single-chain variable fragments, or antibodies
false
Dataset composition
The database includes multiple specialized datasets with diverse characteristics:
RMNA: Binary binding data for 10 antibodies against 2 antigen targets (RiVax and Apolipoprotein A1),
BUZZ: Affinity data for Trastuzumab mutations binding to HER2 (524,346 entries)
DLGO: Examines antibody mutations affecting binding against COVID-19 variants
AAPT: KD affinity measurements for HER2 VHH (nanobody) mutations.
AAE: KD binding affinities for VH antibody variants binding to hen egg lysozyme
AntiBinder: Binary binding data from multiple antibody-antigen pairs.
COVID-19: Neutralization data for 27,324 SARS-CoV-2 antigen–antibody pairs with full VH/VL sequences (Cov-AbDab).
HIV: Heavy/light chain antibody sequences targeting HIV (24,004 pairs from LANL database).
BioMap: Binding ΔG values for 1,706 antigen–antibody complexes across 8 species.
MET: Affinity data for 4,000 emibetuzumab variants targeting the MET receptor.
FLAB: Aggregated data from five publications:
FLAB_Hie2022 (55 datapoints): Binding data for antibodies targeting viral glycoproteins including Ebolavirus glycoprotein, Influenza A hemagglutinin (HA), and its Group A subtype.
FLAB_Koenig2017 (4,275 datapoints): Extensive binding data for antibodies targeting growth factors such as Vascular Endothelial Growth Factor (VEGF).
FLAB_Rosace2023 (25 datapoints): Binding data focused on antibodies targeting Tumor Necrosis Factor-alpha (TNF-α) and SARS-CoV spike proteins.
FLAB_Shanehsazzadeh2023 (445 datapoints): Binding data for antibodies targeting the HER2 receptor, including multiple and zero mutation variants.
FLAB_Warszawski2019 (2,048 datapoints): Binding data associated with antibodies targeting Hen egg lyzosome.
Patent databases: Paired antibody-antigen sequences extracted using NLP techniques (113,122 entries spanning 5291 antigens).
Structure dataset: BBinders extracted from structures in the PDB containing 3969 antigen-antibody pairs.
ABDesign: Systematic point mutations at binding residues of CDR-H3 (1389 single-point mutants).
Literature: Semi-manual dataset containing antibodies and antigens extracted from selected research articles containing 5595 curated unique pairs, including 1940 nanobodies.
AlphaSeq: Dataset involving mutations of antibodies to achieve better binding characteristics across 4 different targets: Human TIGIT, SARS-CoV2_RBD, Human PD-1 and Human HER2 .
OSH: Affinity binding data using Human NKp30 as a target.
ABBD: This dataset offers eight antibody-antigen cases enriched through heavy chain mutations.
Genbank: This dataset offers 413 antigen-antibody pairs derived from the previously published Genbank dataset.
Inclusion criteria
Transparency and completeness of data
Relevance to human health
Quantitative binding affinity measurements
Complete amino acid sequences for all biomolecules
Accessing the Database
AgAb DB is freely available for non-commercial organisations for non-commercial research. Commercial inquiries are welcome via contact us.
It can be accessed in a manner described in the colab notebook.
Citing this work
We make the Therapeutic Antibody Database available as a companion to our paper, that will be posted here once it clears peer review.
Antigen Specific Antibody Database